About Me
Cytoskeletal biologist using comparative cell biology and biochemical reconstitution to understand fundamental principles and novel modes of intracellular transport.
Research
Across the tree of life, cells display remarkable diversity in traits including cellular organization, gene expression, and morphology, among others. These adaptations are critical to the survival of a cell or organism within its unique environment. To perform fundamental cellular processes, each cell must be able to transport cargos such as organelles, vesicles, and mRNAs with spatial and temporal precision. Despite understanding many of the players involved in intracellular transport, little is known about why some organisms use one mode of transport over another. My research vision is to determine the fundamental principles behind how and when cells use different modes of cargo transport. By studying organisms across the tree of life, we can both parse the underlying themes and potentially unearth new modes of intracellular transport.
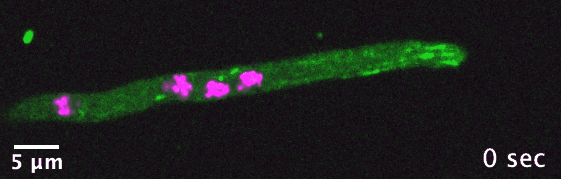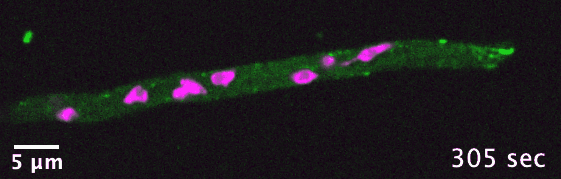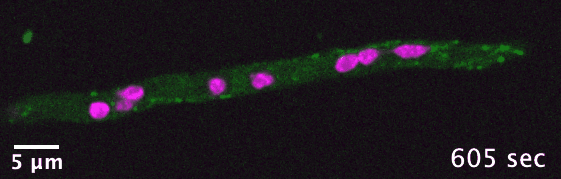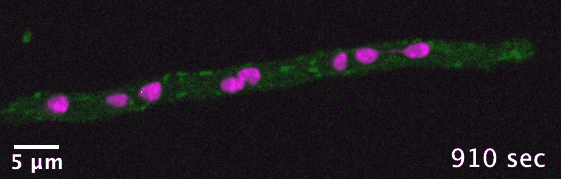 The dikaryotic fungi are an ideal group of organisms in which to study intracellular transport diversification as they exhibit a wide range
of phenotypic diversity, include species known to use actin- and/or microtubule-dependent transport, and encompass many species that are
already genetically tractable. I use comparative genomics to identify transport-related genes encoded in the genomes of different dikaryotic fungi
in order to generate hypotheses concerning intracellular transport adaptation in different contexts.
My research has found that different fungi have different combinations of transport-related genes encoded in their genomes, suggesting that intracellular transport may be fine-tuned in conjunction with cellular
characteristics present in distinct fungal clades. I am now using comparative cell biology in different fungal species to directly test these hypotheses.
The dikaryotic fungi are an ideal group of organisms in which to study intracellular transport diversification as they exhibit a wide range
of phenotypic diversity, include species known to use actin- and/or microtubule-dependent transport, and encompass many species that are
already genetically tractable. I use comparative genomics to identify transport-related genes encoded in the genomes of different dikaryotic fungi
in order to generate hypotheses concerning intracellular transport adaptation in different contexts.
My research has found that different fungi have different combinations of transport-related genes encoded in their genomes, suggesting that intracellular transport may be fine-tuned in conjunction with cellular
characteristics present in distinct fungal clades. I am now using comparative cell biology in different fungal species to directly test these hypotheses.
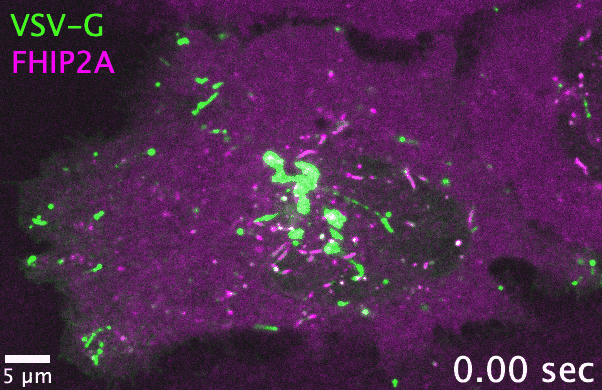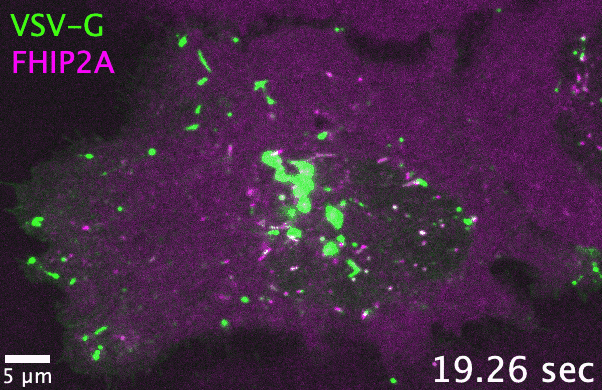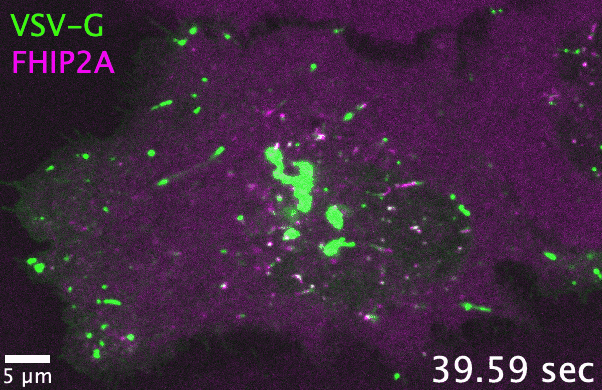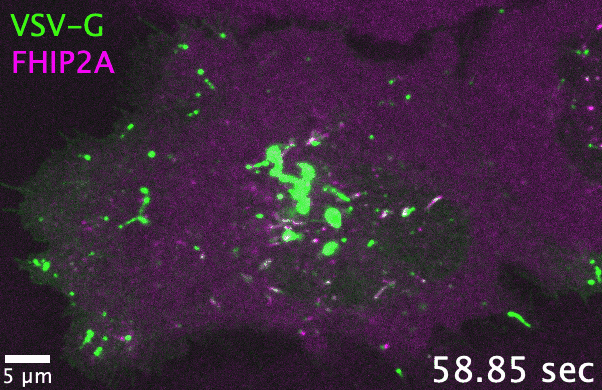 Unlike filamentous fungi, in which cellular cargos are often distributed consistently throughout a hyphal network, cargo transport in human cells
must be multi-dimensional, comprehensive, and responsive. Correspondingly, human cells have evolved a large series of functionally distinct motors
and adaptors. As one example, the filamentous fungus Aspergillus nidulans has one each of the FTS, Hook, and FHIP proteins, which collectively
form the ‘FHF’ complex and link dynein to early endosomes. In comparison, human cells have three Hook, four FHIP proteins, and one FTS. I discovered
that gene family expansion and functional divergence of FHF complex proteins allow dynein to bind multiple cargos in human cells. Many transport-related
gene families have similarly expanded in humans. My future work uses fungi as models to investigate general roles and mechanisms of transport-related proteins,
and then uses cellular proteomics and cell biology to discover how those processes have diverged or expanded in human cells.
Unlike filamentous fungi, in which cellular cargos are often distributed consistently throughout a hyphal network, cargo transport in human cells
must be multi-dimensional, comprehensive, and responsive. Correspondingly, human cells have evolved a large series of functionally distinct motors
and adaptors. As one example, the filamentous fungus Aspergillus nidulans has one each of the FTS, Hook, and FHIP proteins, which collectively
form the ‘FHF’ complex and link dynein to early endosomes. In comparison, human cells have three Hook, four FHIP proteins, and one FTS. I discovered
that gene family expansion and functional divergence of FHF complex proteins allow dynein to bind multiple cargos in human cells. Many transport-related
gene families have similarly expanded in humans. My future work uses fungi as models to investigate general roles and mechanisms of transport-related proteins,
and then uses cellular proteomics and cell biology to discover how those processes have diverged or expanded in human cells.
 Factors such as cytoskeletal organization, motor composition, and post-translational modifications can affect intracellular transport on the molecular level.
By reconstituting different transport machineries and visualizing their activity via single molecule TIRF microscopy, we can elucidate the roles that these factors and others play
in the transport of cargos in different organisms.
Factors such as cytoskeletal organization, motor composition, and post-translational modifications can affect intracellular transport on the molecular level.
By reconstituting different transport machineries and visualizing their activity via single molecule TIRF microscopy, we can elucidate the roles that these factors and others play
in the transport of cargos in different organisms.
Education
Self-organization of the fission yeast actin cytoskeleton
Research & Work Experiences
- Identified novel regulators of hitchhiking, a non-canonical form of transport in Aspergillus nidulans (read more here)
- Discovered that combinatorial assembly of ‘FHF’ dynein adaptor complexes link dynein to different cargos in human cells (read more here)
- Used comparative genomics analysis to discover novel modes of nuclear transport in Aspergillus nidulans (current unpublished work).
- Performed key cell biology experiments and intellectual contribution to build mathematical models defining parameters that affect cargo transport (read more here and here.)
- Significant mentorship of 2 PhD and 4 Undergraduate students.
- Developed complex in vitro reconstitution assays of actin cytoskeleton networks in conjunction with four-color single molecule TIRF microscopy (read more here).
- Discovered that actin binding proteins compete by different mechanisms that are critical for proper sorting in fission yeast (read more here and here.)
- Found that actin binding proteins formin and profilin are tailored to generate actin assembly at the correct time and place in fission yeast and Chlamydomonas reinhardtii (read more here and here.)
